

This neglected tropical disease has a close relationship with poverty, being a major challenge to public health in countries where it remains endemic. It still remains a stigmatizing disease (Nascimento 2013 Degang et al. The disease displays a spectrum of clinical manifestations, such as lepromatous (multibacillar) and tuberculoid (paucibacillar) leprosy, which are attributed to the host immune response. The skin, the peripheral nerves, the nasal mucosa, eyes, and the reticulum-endothelial system are the preferred target sites for this pathogen. Mycobacterium leprae is the causative agent of leprosy, an ancient chronic infectious disease and may have severely debilitating physical, social, and psychological consequences.
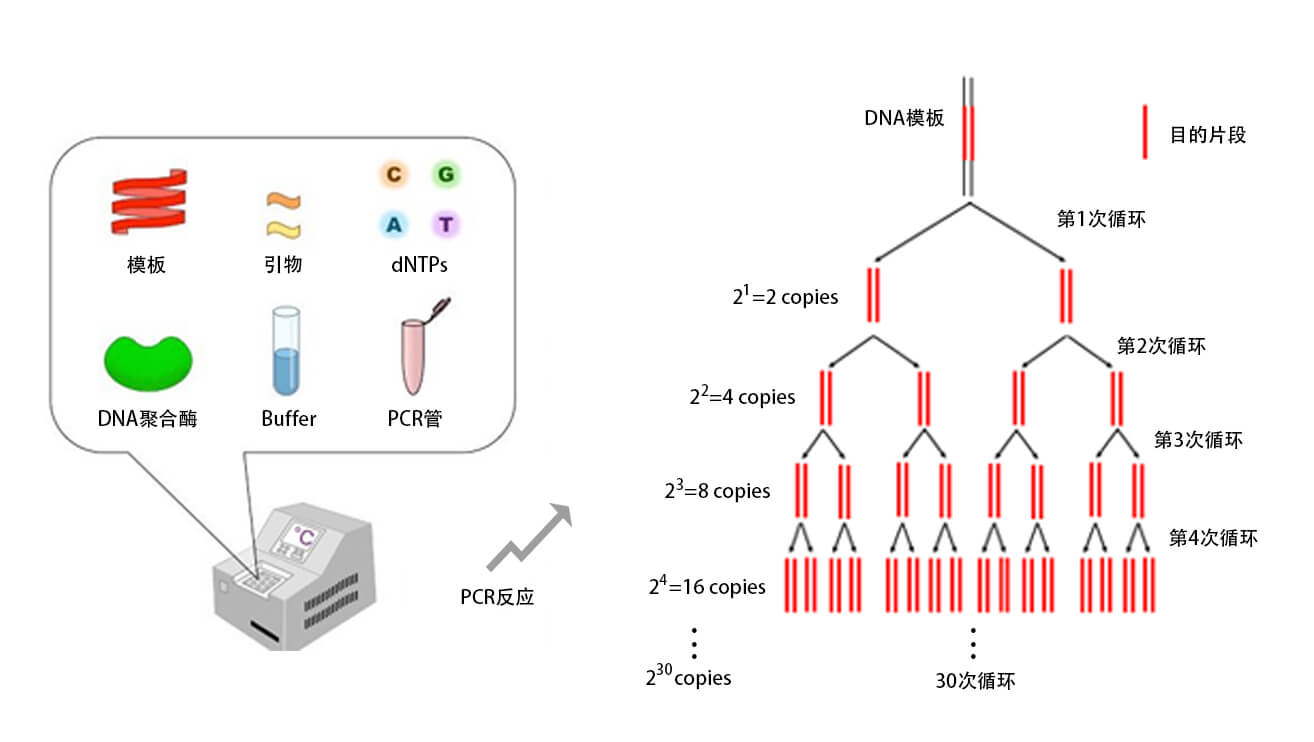
#Touchdown pcr microbiome skin
Our data suggest significant shifts of the microbiota with emergence and competitive advantage of potentially pathogenic bacteria over skin resident taxa. Most of the taxa found in skin from leprous lesions are not typical in human skin and potentially pathogenic, with the Burkholderia, Pseudomonas and Bacillus genera being overrepresented. Propionibacterium, Corynebacterium and Staphylococcus, resident and abundant in healthy skin, were underrepresented in skin from leprous lesions.
There were considerable differences in the distribution of Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria, with the first two phyla enriched and the other markedly diminished in the leprous lesions, when compared with healthy skin. Taxonomic analysis of leprous lesions revealed main four phyla: Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria, with Proteobacteria presenting the highest diversity. Here we used Sanger and massively parallel small sub-unit rRNA (SSU) rRNA gene sequencing to characterize the microbiota of leprous lesions, and studied how it differs from the bacterial skin composition of healthy individuals previously described in the literature. But while studies are continuously revealing the complexity of human skin microbiota, the microbiota of leprous cutaneous lesions has not yet been characterized. Increasing evidence has highlighted the importance of microbiota for human general health and, as such, the study of skin microbiota is of interest. Some commensal coagulase-negative bacteria may have a role as mecA resistance reservoirs.Leprosy is a chronic infectious disease that remains a major challenge to public health in endemic countries. Moreover, we detected a new putative variant of this gene. This is the first report of blaOXA-181 in environmental samples in Ecuador. β-lactam antibiotic resistance genes are prevalent throughout QPT. Almost half of the circulating bacteria found at QPT stations were common human microbiota species, including those classified by the WHO as pathogens of critical and high-priority surveillance. Two subvariants were found for blaTEM-1, blaCTX-M-1, and blaOXA-181. ESBL genes blaTEM-1 and blaCTX-M-1 and MBL genes blaOXA-181 and mecA were detected along QPT stations, blaTEM being the most widely spread. PCRs were performed to detect the presence of 13 antibiotic resistance genes and to identify and to amplify 16S rDNA for barcoding, followed by clone analysis, Sanger sequencing, and BLAST search. A total of 29 station turnstiles were swabbed to extract the surface environmental DNA. This study aimed to determine the presence and spread of β-lactam antibiotic resistance genes and the microbiome circulating in Quito's Public Transport (QTP). Likewise, Gram-positive bacteria have evolved other mechanisms through mec genes, which encode modified penicillin-binding proteins (PBP2). Multidrug-resistant bacteria present resistance mechanisms against β-lactam antibiotics, such as Extended-Spectrum Beta-lactamases (ESBL) and Metallo-β-lactamases enzymes (MBLs) which are operon encoded in Gram-negative species. Int J Environ Res Public Health (IF: 4.6) 20(3) Abstract Hernández-Alomía, Bastidas-Caldes, Ballesteros, Tenea, Jarrín-V, Molina, Castillejo (2023) Beta-Lactam Antibiotic Resistance Genes in the Microbiome of the Public Transport System of Quito, Ecuador. Beta-Lactam Antibiotic Resistance Genes in the Microbiome of the Public Transport System of Quito, Ecuador.


 0 kommentar(er)
0 kommentar(er)
